Creates a Venn diagram comparing the number of common sequences in two or three repertoire_ids using native ggplot2 implementation.
Arguments
- repertoire_ids
A character vector of two or three names of repertoire_ids in
productiveSeq()table to compare.- amino_table
A tibble of amino acid sequences generated by the function
productiveSeq().
Value
Returns a ggplot2 Venn diagram of the number of common sequences between two or three repertoire_ids.
Examples
file_path <- system.file("extdata", "TCRB_sequencing",
package = "LymphoSeq2")
study_table <- LymphoSeq2::readImmunoSeq(path = file_path, threads = 1)
#> Dataset Analysis:
#> Files: 10, Total: 0.00 GB, Largest: 0.0 MB
#> Available memory: 12.0 GB
study_table <- LymphoSeq2::topSeqs(study_table, top = 100)
amino_table <- LymphoSeq2::productiveSeq(
study_table = study_table,
aggregate = "junction_aa"
)
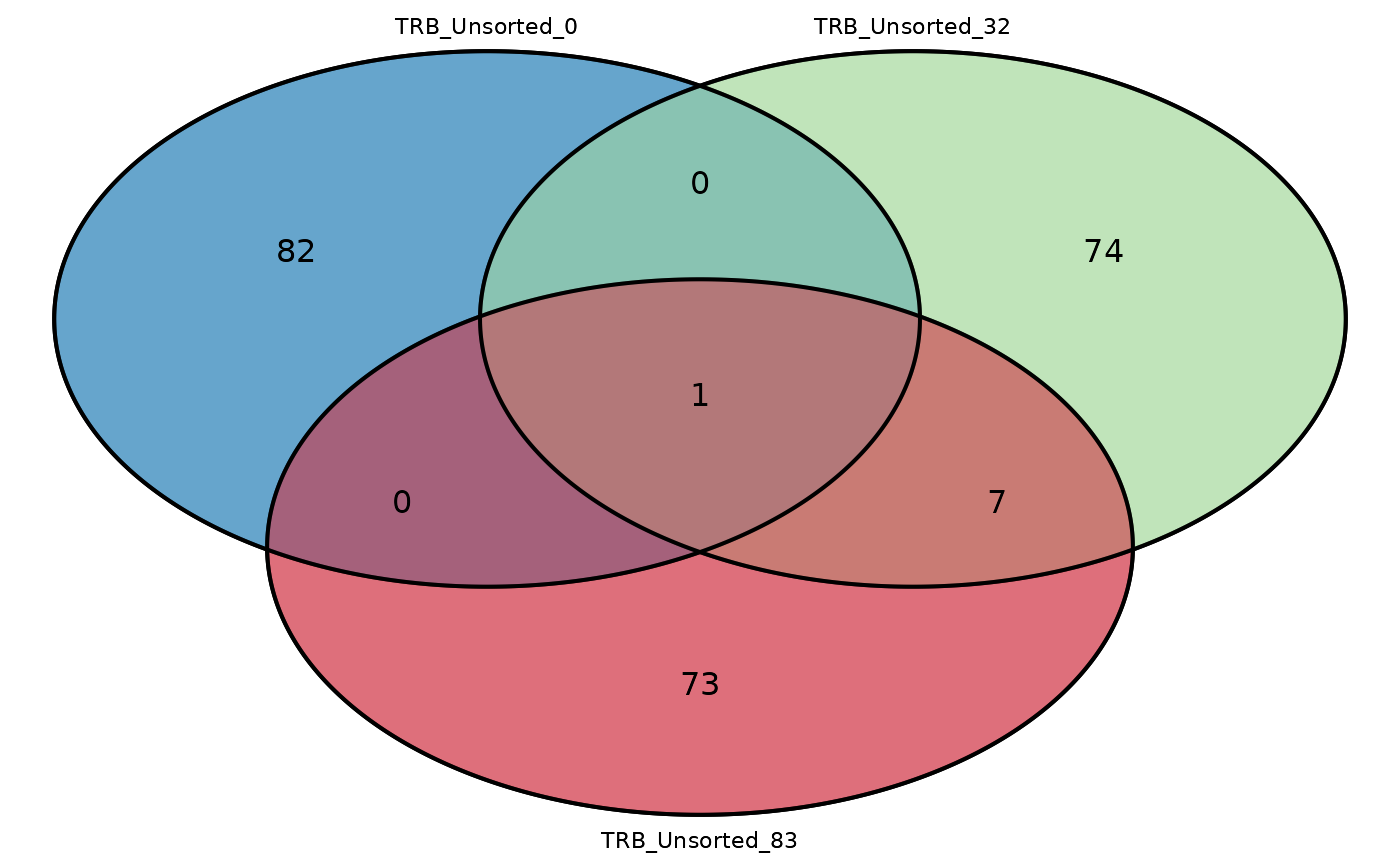
# Plot a triple Venn diagram
LymphoSeq2::commonSeqsVenn(
repertoire_ids = c(
"TRB_Unsorted_0",
"TRB_Unsorted_32", "TRB_Unsorted_83"
),
amino_table = amino_table
)
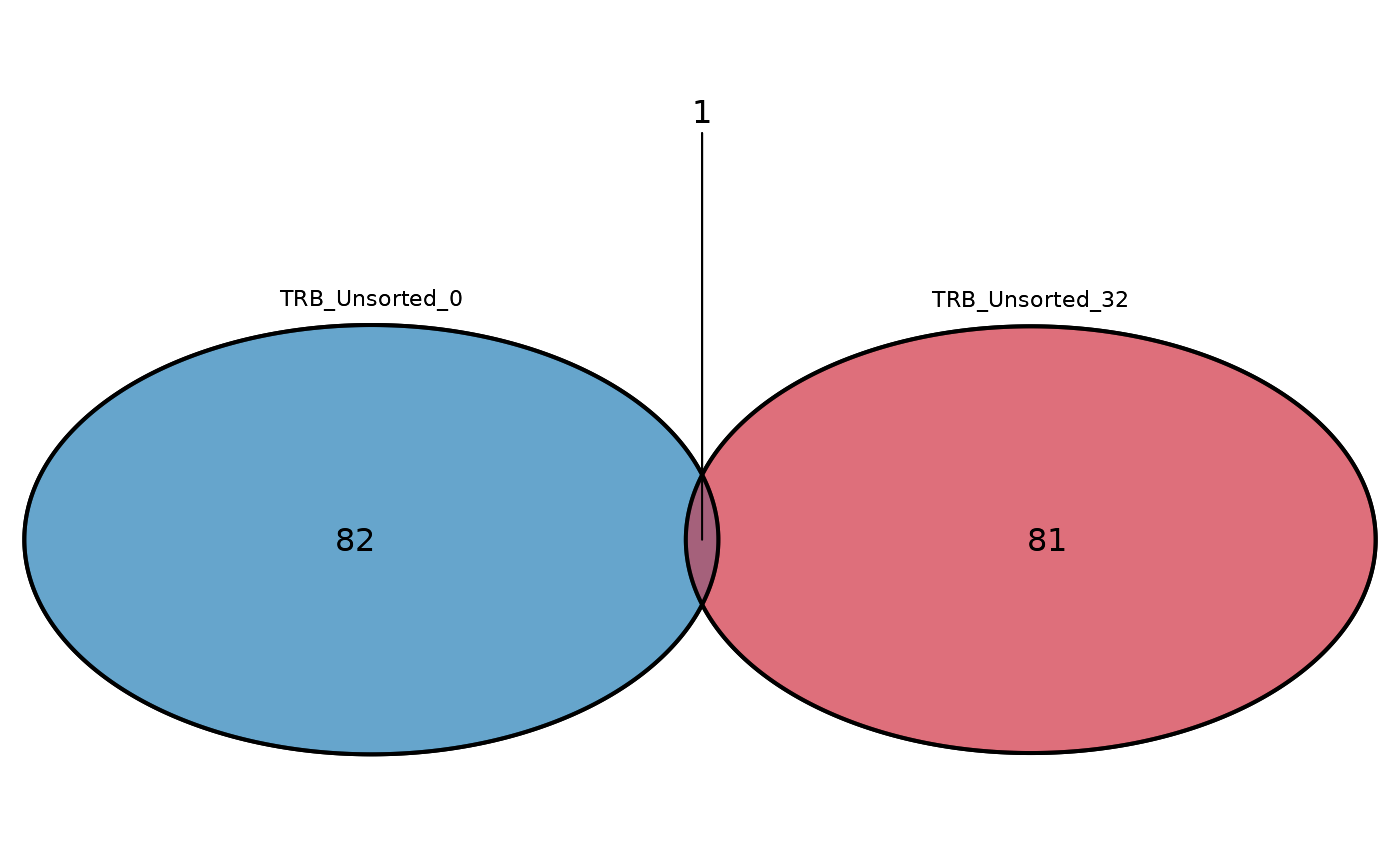
 # Plot a double Venn diagram
LymphoSeq2::commonSeqsVenn(repertoire_ids = c(
"TRB_Unsorted_0",
"TRB_Unsorted_32"
), amino_table = amino_table)
# Plot a double Venn diagram
LymphoSeq2::commonSeqsVenn(repertoire_ids = c(
"TRB_Unsorted_0",
"TRB_Unsorted_32"
), amino_table = amino_table)